3 The Central Dogma
3.1 The Most Important Rule in Biology
3.1.1 What Is the Central Dogma?
The Central Dogma is the most important rule about how genetic information flows in living things. It describes the journey from DNA to proteins.
Don’t worry—“dogma” is just a fancy word for a rule or principle. And “central” means it’s super important!
3.1.2 The Simple Version
Here’s the central dogma in its simplest form:
DNA → RNA → Protein
Or in words: DNA makes RNA, and RNA makes Protein
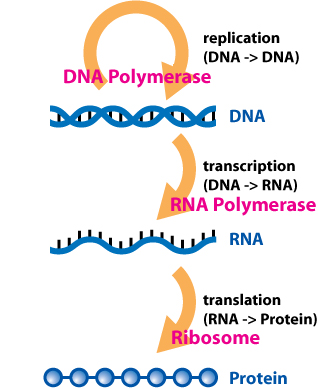
Figure 3.1: The Central Dogma of molecular biology showing the flow of genetic information from DNA to RNA (transcription) to Protein (translation).
Image credit: Wikimedia Commons, CC BY-SA 4.0
Think of it like a factory assembly line:
DNA is the blueprint/instruction manual (stored in the office)
RNA is the work order/copy of instructions (delivered to the factory floor)
Protein is the finished product (built following the instructions)
3.2 The Two Main Steps
The central dogma has two main steps with fancy names. Let’s learn what they mean!
3.2.1 Step 1: Transcription (DNA → RNA)
Transcription means “copying” or “writing out.”
3.2.1.1 What Happens:
The DNA stays safely in the nucleus (like the original cookbook stays in the library)
A special enzyme “reads” the DNA
It makes a copy of the instructions using RNA letters
This copy is called mRNA (messenger RNA)
3.2.1.2 Think of It Like This:
Imagine you have a precious, old cookbook you want to protect:
You wouldn’t take it into the kitchen where it might get dirty
Instead, you’d photocopy the recipe you need
The photocopy goes to the kitchen, not the original book!
That’s exactly what transcription does—it makes a temporary copy (RNA) of the permanent instructions (DNA).
3.2.1.3 The Details:
DNA unzips (the twisted ladder opens up)
An enzyme called RNA polymerase reads one side of the DNA
It builds an RNA strand using the DNA as a template
The RNA uses the same letters as DNA, except U (Uracil) instead of T (Thymine)
Pairing Rules:
DNA has A → RNA gets U
DNA has T → RNA gets A
DNA has G → RNA gets C
DNA has C → RNA gets G
3.2.2 Step 2: Translation (RNA → Protein)
Translation means changing from one language to another.
3.2.2.1 What Happens:
The mRNA leaves the nucleus and goes to a protein-making factory called a ribosome
The ribosome “reads” the RNA code
It translates the RNA letters into amino acids
The amino acids link together to make a protein
3.2.2.2 Think of It Like This:
Imagine you have a recipe written in French, but you speak English:
You need to translate the recipe from French to English
Once translated, you can follow the instructions to make the dish
That’s what translation does—it changes the “language” from RNA code into amino acid sequences!
3.2.2.3 The Details:
mRNA attaches to a ribosome
The ribosome reads the mRNA in groups of 3 letters at a time
Each group of 3 letters is called a codon
Each codon tells the ribosome which amino acid to add next
tRNA (transfer RNA) brings the correct amino acids
The amino acids link together like beads on a string to form a protein
3.2.2.4 The Genetic Code:
Every 3 letters of RNA code for 1 amino acid. For example:
AUG = Start making the protein! (and codes for amino acid Methionine)
UUU = Add the amino acid Phenylalanine
GGG = Add the amino acid Glycine
UAA = Stop! The protein is finished!
This code is used by almost all living things on Earth—from bacteria to blue whales to you!
3.3 Replication: Making Copies of DNA
Before we move on, there’s one more important process: Replication.
3.3.1 What Is Replication?
Replication means making a copy of DNA.
Every time a cell divides (to make new cells), it needs to copy all its DNA so the new cell gets a complete set of instructions.
3.3.1.1 How It Works:
The DNA double helix unzips down the middle
Each side serves as a template
New DNA letters attach to each side following the pairing rules (A with T, G with C)
You end up with two identical copies of the original DNA
Figure 3.2: Semi-conservative DNA replication showing the double helix unwinding and each strand serving as a template for a new complementary strand.
Image credit: Mariana Ruiz LadyofHats, Wikimedia Commons, CC BY-SA 3.0
Think of it like:
Unzipping a zipper
Each side of the zipper gets new matching teeth
Now you have two complete zippers!
3.4 Exceptions to the Rule
The central dogma works for most living things, most of the time. But biology has some rule-breakers!
3.4.1 Exception 1: Reverse Transcription
What Normally Happens: DNA → RNA → Protein
What Sometimes Happens: RNA → DNA
Some viruses (like HIV) store their genetic information as RNA instead of DNA. When they infect a cell, they can make DNA from their RNA using an enzyme called reverse transcriptase.
It’s like going backward on a one-way street!
3.4.2 Exception 2: RNA-Dependent RNA Synthesis
What Normally Happens: DNA → RNA → Protein
What Sometimes Happens: RNA → RNA
Some viruses can make RNA directly from RNA, without using DNA at all!
3.4.3 Exception 3: Proteins Directly Affecting DNA
Sometimes proteins can affect which genes are turned on or off, creating a feedback loop. It’s not breaking the central dogma, but it shows that biology is more complex than a simple one-way street!
3.6 Putting It All Together
Let’s review the complete journey from DNA to protein:
3.6.1 The Normal Path:
Replication (when cells divide)
DNA → DNA
Making copies of the instruction manual
Transcription (making mRNA)
DNA → RNA
Copying instructions from the manual
Translation (making proteins)
RNA → Protein
Building the final product
3.6.2 The Big Picture:
Think of your cells like a factory:
DNA = The master blueprints (kept safe in the office)
Transcription = Photocopying specific blueprints
mRNA = The photocopied work orders
Translation = Following the work orders on the factory floor
Proteins = The finished products that do the work
Replication = Making exact copies of all the blueprints (for new factories)
3.7 Why Is This Important?
Understanding the central dogma helps us:
Understand Diseases
Many diseases happen when something goes wrong in these processes
Some medicines work by affecting transcription or translation
Develop New Technologies
Scientists can now “edit” DNA (like spell-check for genes!)
We can make bacteria produce human proteins (like insulin for diabetes)
Study Evolution
All living things use the same basic process
This shows we’re all connected through deep time
Personalize Medicine
- Understanding your DNA can help doctors choose the best treatments for you
3.8 Codon Bias: Not All Codons Are Equal!
3.8.1 The Redundancy Problem
Remember the genetic code is redundant (also called “degenerate”):
64 possible codons (4³ = 4 × 4 × 4)
Only 20 amino acids (+ 3 stop signals)
Multiple codons code for the same amino acid!
Example - Leucine:
Has 6 different codons: UUA, UUG, CUU, CUC, CUA, CUG
All mean “add leucine”
So which one does the cell use?
You might think: All codons for an amino acid used equally, right?
Wrong! Different organisms have codon preferences (codon bias)!
3.8.2 What Is Codon Bias?
Codon bias = Organisms preferentially use certain codons over others, even though they code for the same amino acid
Example in E. coli bacteria:
Leucine codon CTG is used very often
Leucine codon CTA is used rarely
Both code for leucine!
But bacteria “prefer” CTG
Why does this happen?
1. tRNA Availability:
Different organisms have different amounts of different tRNAs
Preferred codons match abundant tRNAs
Allows faster, more efficient translation
2. Translation Efficiency:
Highly expressed genes use “optimal” codons
Optimal = codons with abundant matching tRNAs
Faster translation = more protein made
3. Translation Accuracy:
Some codons translated more accurately
Important genes use more accurate codons
Reduces errors in critical proteins
4. Evolutionary Selection:
Natural selection favors efficient codon usage
Especially in fast-growing organisms (like bacteria)
Less pressure in slow-growing organisms
3.8.3 Codon Bias Is Organism-Specific!
Different organisms have different codon preferences!
Examples:
| Amino Acid | Humans Prefer | E. coli Prefers | Yeast Prefers |
|---|---|---|---|
| Leucine | CTG | CTG | TTG |
| Arginine | CGC | CGT | AGA |
| Serine | AGC | AGC | TCT |
This matters a LOT for biotechnology!
3.8.4 Practical Problem: Heterologous Gene Expression
“Heterologous expression” = Making one organism produce a protein from another organism
Common scenario:
Take human gene
Put it in bacteria
Make bacteria produce human protein (like insulin!)
The problem:
Human gene uses human codon preferences
Bacteria have different tRNA abundances
→ Bacteria struggle to translate human gene!
→ Low protein production or even failure!
**Real example - Human insulin production**:
- Original human insulin gene → poor expression in E. coli
- Many codons rare in bacteria
- Translation stalls, protein misfolds
- **Solution needed!**
### Solution: Codon Optimization
**Codon optimization** = Changing DNA sequence to use host organism's preferred codons, **without changing the amino acid sequence**!
**How it works**:
1. Take human gene sequence
2. Identify which codons are rare in bacteria
3. Replace with synonymous codons preferred by bacteria
4. **Amino acid sequence stays identical!**
5. But bacteria translate it much better!
**Example - Leucine in human vs bacteria**:Original human gene: ...UUA-UUA-UUA... (rare in bacteria)
↓
Optimized for E. coli: ...CUG-CUG-CUG... (common in bacteria)
Both sequences code for: Leu-Leu-Leu
But bacteria prefer CUG!
**Results after optimization**:
- ✅ 10-100x more protein produced!
- ✅ Better protein folding
- ✅ Higher purity
- ✅ More economical production
**Real-world applications**:
- Insulin for diabetes (optimized for E. coli)
- Vaccines (optimized for yeast or bacteria)
- Industrial enzymes
- Research proteins
- Antibody production
### Codon Usage in Gene Prediction
Codon bias also helps in **bioinformatics**!
**How computers find genes**:
- Calculate codon usage in DNA sequence
- Compare to organism's known codon preferences
- Regions matching codon bias → likely coding!
- Regions not matching → likely non-coding
**Example**:Sequence A: Uses organism's preferred codons → Probably a gene!
Sequence B: Random codon usage → Probably not a gene```
This is one signal used in ab initio gene prediction (covered in Chapter 17a)!
3.8.5 Codon Adaptation Index (CAI)
CAI = Measure of how well a gene’s codons match organism’s preferences
Scale: 0 to 1
CAI = 1.0: Perfect match (uses all optimal codons)
CAI = 0.5: Medium match
CAI < 0.3: Poor match (uses rare codons)
In practice:
Highly expressed genes: CAI ~0.7-0.9
Lowly expressed genes: CAI ~0.3-0.5
Foreign genes (before optimization): CAI often < 0.3
Use in biotech:
Calculate CAI before cloning gene
If CAI < 0.4 in host → optimize!
Predicts expression success
3.8.6 Exceptions and Special Cases
Not all organisms show strong codon bias:
Bacteria: Strong bias (need efficiency!)
Yeast: Moderate bias
Mammals: Weaker bias (can afford inefficiency)
Plants: Variable bias
Why weaker bias in complex organisms?
Slower growth rate (less time pressure)
More complex regulation needed
Different selection pressures
Rare codons can be functional!
Sometimes rare codons used intentionally
Slow down translation at specific points
Allow proper protein folding
Regulatory mechanism!
3.8.7 Key Takeaways - Codon Bias
Redundant genetic code: Multiple codons per amino acid
Codon bias: Organisms prefer certain synonymous codons
Organism-specific: Each species has different preferences
Linked to tRNA availability: Preferred codons have abundant tRNAs
Affects translation efficiency: Optimal codons = faster translation
Critical for biotech: Must optimize codons for heterologous expression
Used in gene prediction: Codon usage helps identify coding regions
CAI metric: Measures codon optimization
Can be regulatory: Rare codons sometimes serve functions
Bottom line: The genetic code is universal, but codon preferences are not!
3.9 Fun Facts! 🎉
Your cells transcribe genes into RNA constantly—thousands of times per second!
A typical human cell makes about 10,000 different proteins
Translation happens incredibly fast—a ribosome can add about 20 amino acids per second
The genetic code is almost universal—the same codons mean the same amino acids in nearly all living things
Some proteins are just a few dozen amino acids long, while others have thousands!
Bacteria can prefer certain codons so strongly that rare codons are used <5% as often as optimal ones!
3.10 Key Takeaways
Central Dogma: DNA → RNA → Protein (the flow of genetic information)
Transcription: Making RNA from DNA (like photocopying a recipe)
Translation: Making proteins from RNA (like following the recipe to cook)
Replication: Making copies of DNA (when cells divide)
Genetic Code: 3 RNA letters = 1 amino acid
Exceptions exist: Some viruses use reverse transcription (RNA → DNA)
Universal process: Almost all living things use the same basic system
This understanding helps us develop medicines, study evolution, and understand diseases
Sources: Information adapted from Khan Academy (Intro to Gene Expression - Central Dogma), NHGRI Genetics Glossary, Nature Scitable, and Biology LibreTexts (Central Dogma of Molecular Biology).
This is one signal used in ab initio gene prediction (covered in Chapter 17a)!
3.10.1 Codon Adaptation Index (CAI)
CAI = Measure of how well a gene’s codons match organism’s preferences
Scale: 0 to 1
CAI = 1.0: Perfect match (uses all optimal codons)
CAI = 0.5: Medium match
CAI < 0.3: Poor match (uses rare codons)
In practice:
Highly expressed genes: CAI ~0.7-0.9
Lowly expressed genes: CAI ~0.3-0.5
Foreign genes (before optimization): CAI often < 0.3
Use in biotech:
Calculate CAI before cloning gene
If CAI < 0.4 in host → optimize!
Predicts expression success
3.10.2 Exceptions and Special Cases
Not all organisms show strong codon bias:
Bacteria: Strong bias (need efficiency!)
Yeast: Moderate bias
Mammals: Weaker bias (can afford inefficiency)
Plants: Variable bias
Why weaker bias in complex organisms?
Slower growth rate (less time pressure)
More complex regulation needed
Different selection pressures
Rare codons can be functional!
Sometimes rare codons used intentionally
Slow down translation at specific points
Allow proper protein folding
Regulatory mechanism!
3.10.3 Key Takeaways - Codon Bias
Redundant genetic code: Multiple codons per amino acid
Codon bias: Organisms prefer certain synonymous codons
Organism-specific: Each species has different preferences
Linked to tRNA availability: Preferred codons have abundant tRNAs
Affects translation efficiency: Optimal codons = faster translation
Critical for biotech: Must optimize codons for heterologous expression
Used in gene prediction: Codon usage helps identify coding regions
CAI metric: Measures codon optimization
Can be regulatory: Rare codons sometimes serve functions
Bottom line: The genetic code is universal, but codon preferences are not!
3.11 Fun Facts! 🎉
Your cells transcribe genes into RNA constantly—thousands of times per second!
A typical human cell makes about 10,000 different proteins
Translation happens incredibly fast—a ribosome can add about 20 amino acids per second
The genetic code is almost universal—the same codons mean the same amino acids in nearly all living things
Some proteins are just a few dozen amino acids long, while others have thousands!
Bacteria can prefer certain codons so strongly that rare codons are used <5% as often as optimal ones!
3.12 Key Takeaways
Central Dogma: DNA → RNA → Protein (the flow of genetic information)
Transcription: Making RNA from DNA (like photocopying a recipe)
Translation: Making proteins from RNA (like following the recipe to cook)
Replication: Making copies of DNA (when cells divide)
Genetic Code: 3 RNA letters = 1 amino acid
Exceptions exist: Some viruses use reverse transcription (RNA → DNA)
Universal process: Almost all living things use the same basic system
This understanding helps us develop medicines, study evolution, and understand diseases
Sources: Information adapted from Khan Academy (Intro to Gene Expression - Central Dogma), NHGRI Genetics Glossary, Nature Scitable, and Biology LibreTexts (Central Dogma of Molecular Biology).